Reading data from the web
In data import, we saw how to load data from a variety of formats; this is a fairly standard way to get data that have been gathered as part of a study. In a lot of cases, though, you’re going to have to go out and get the data you want or need. That’s what we’re covering now.
This is the first module in the Data Wrangling II topic; the relevant slack channel is here.
Example
As always, it’s good to start by figuring out how you want to organize you code for this content. I’ll create a new directory called example_data_wrangling_ii, start an R Project, and open a new R Markdown file called reading_data_from_the_web.Rmd. I’ll initialize Git and make the repo public on GitHub. Although we’ll mostly be getting data from the web, we’ll revisit some of these examples, so I’ll create a data subdirectory and put those in it.
There are some new additions to our standard packages (rvest and httr); I’m loading everything we need now. Now’s also the time to “install” the Selector Gadget.
library(tidyverse)
library(rvest)
## Loading required package: xml2
##
## Attaching package: 'rvest'
## The following object is masked from 'package:purrr':
##
## pluck
## The following object is masked from 'package:readr':
##
## guess_encoding
library(httr)
##
## Attaching package: 'httr'
## The following object is masked from 'package:plotly':
##
## configExtracting tables
This page contains data from the National Survey on Drug Use and Health; it includes tables for drug use in the past year or month, separately for specific kinds of drug use. These data are potentially useful for analysis, and we’d like to be able to read in the first table.
First, let’s make sure we can load the data from the web.
url = "http://samhda.s3-us-gov-west-1.amazonaws.com/s3fs-public/field-uploads/2k15StateFiles/NSDUHsaeShortTermCHG2015.htm"
drug_use_xml = read_html(url)
drug_use_xml
## {xml_document}
## <html lang="en">
## [1] <head>\n<link rel="P3Pv1" href="http://www.samhsa.gov/w3c/p3p.xml">\ ...
## [2] <body>\r\n\r\n<noscript>\r\n<p>Your browser's Javascript is off. Hyp ...Doesn’t look like much, but we’re there. Rather than trying to grab something using a CSS selector, let’s try our luck extracting the tables from the HTML.
drug_use_xml %>%
html_nodes(css = "table")
## {xml_nodeset (15)}
## [1] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [2] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [3] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [4] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [5] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [6] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [7] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [8] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [9] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [10] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [11] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [12] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [13] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [14] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...
## [15] <table class="rti" border="1" cellspacing="0" cellpadding="1" width ...This has extracted all of the tables on the original page; that’s why we have a list with 15 elements. We’re only focused on the first one for now, so let’s get the contents from the first list element.
table_marj = (drug_use_xml %>% html_nodes(css = "table"))[[1]] %>%
html_table() I won’t print the table here, but if you look at it you’ll notice a problem: the “note” at the bottom of the table appears in every column in the first row. We need to remove that; I’ll also convert to a tibble so that things print nicely.
table_marj = (drug_use_xml %>% html_nodes(css = "table"))[[1]] %>%
html_table() %>%
.[-1,] %>%
as_tibble()
table_marj
## # A tibble: 56 x 16
## State `12+(2013-2014)` `12+(2014-2015)` `12+(P Value)` `12-17(2013-201…
## * <chr> <chr> <chr> <chr> <chr>
## 1 Total… 12.90a 13.36 0.002 13.28b
## 2 North… 13.88a 14.66 0.005 13.98
## 3 Midwe… 12.40b 12.76 0.082 12.45
## 4 South 11.24a 11.64 0.029 12.02
## 5 West 15.27 15.62 0.262 15.53a
## 6 Alaba… 9.98 9.60 0.426 9.90
## # ... with 50 more rows, and 11 more variables: `12-17(2014-2015)` <chr>,
## # `12-17(P Value)` <chr>, `18-25(2013-2014)` <chr>,
## # `18-25(2014-2015)` <chr>, `18-25(P Value)` <chr>,
## # `26+(2013-2014)` <chr>, `26+(2014-2015)` <chr>, `26+(P Value)` <chr>,
## # `18+(2013-2014)` <chr>, `18+(2014-2015)` <chr>, `18+(P Value)` <chr>Success!! At least, mostly. These data aren’t tidy, but we’ll worry about that in another module.
Learning assessment: Create a data frame that contains the cost of living table for New York from this page.
CSS Selectors
Suppose we’d like to scrape the cast of Harry Potter and the Sorcerer’s Stone from the IMDB page. The first step is the same as before – we need to get the HTML.
hpss_html = read_html("http://www.imdb.com/title/tt0241527/")The cast list isn’t stored in a handy table, so we’re going to isolate the CSS selector for cast. A bit of clicking around gets me something like below.
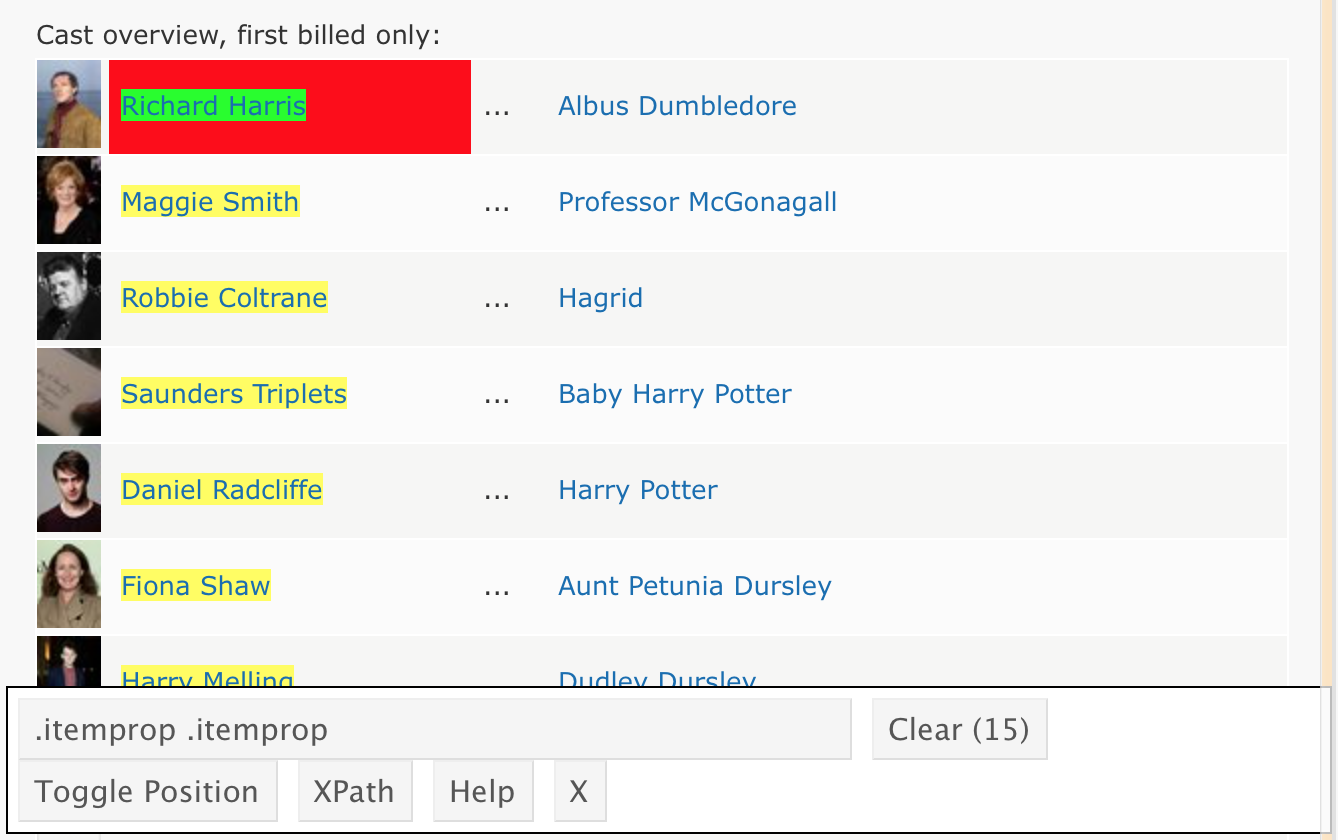
I’ll use the CSS selector in html_nodes() to extract the relevant HTML code, and convert it to text.
cast = hpss_html %>%
html_nodes(".itemprop .itemprop") %>%
html_text()Learning Assessment: This page contains the 10 most recent reviews of a popular toothbrush. Use a process similar to the one above to extract the titles of the reviews. Note: getting the star ratings from Amazon is trickier, but the CSS path "#cm_cr-review_list .review-rating" helps – I discovered this after about an hour of googling around.
Using an API
New York City has a great open data resource, and we’ll use that for our API examples. Although most (all?) of these datasets can be accessed by clicking through a website, we’ll access them directly using the API to improve reproducibility and make it easier to update results to reflect new data.
As a simple example, this page is about a dataset for annual water consumption in NYC, along with the population in that year. First, we’ll import this as a CSV and parse it.
nyc_water = GET("https://data.cityofnewyork.us/resource/waf7-5gvc.csv") %>%
content("parsed")
## Parsed with column specification:
## cols(
## new_york_city_population = col_double(),
## nyc_consumption_million_gallons_per_day = col_double(),
## per_capita_gallons_per_person_per_day = col_integer(),
## year = col_integer()
## )We can also import this dataset as a JSON file. This takes a bit more work (and this is, really, a pretty easy case), but it’s still doable.
nyc_water = GET("https://data.cityofnewyork.us/resource/waf7-5gvc.json") %>%
content("text") %>%
jsonlite::fromJSON() %>%
as_tibble()Data.gov also has a lot of data available using their API; often this is available as CSV or JSON as well. For example, we might be interested in data coming from BRFSS. This is importable via the API as a CSV (JSON, in this example, is much more complicated).
brfss =
GET("https://chronicdata.cdc.gov/api/views/hn4x-zwk7/rows.csv?accessType=DOWNLOAD") %>%
content("parsed")
## Parsed with column specification:
## cols(
## .default = col_character(),
## YearStart = col_integer(),
## YearEnd = col_integer(),
## Data_Value = col_double(),
## Data_Value_Alt = col_double(),
## Low_Confidence_Limit = col_double(),
## High_Confidence_Limit = col_double(),
## Sample_Size = col_integer()
## )
## See spec(...) for full column specifications.Both of the previous examples are extremely easy – we accessed data that is essentially a data table, and we had a very straightforward API.
To get a sense of how this becomes complicated, let’s look at the Pokemon API (which is also pretty nice).
poke = GET("http://pokeapi.co/api/v2/pokemon/1") %>%
content()
poke$name
## [1] "bulbasaur"
poke$height
## [1] 7
poke$abilities
## [[1]]
## [[1]]$slot
## [1] 3
##
## [[1]]$is_hidden
## [1] TRUE
##
## [[1]]$ability
## [[1]]$ability$url
## [1] "https://pokeapi.co/api/v2/ability/34/"
##
## [[1]]$ability$name
## [1] "chlorophyll"
##
##
##
## [[2]]
## [[2]]$slot
## [1] 1
##
## [[2]]$is_hidden
## [1] FALSE
##
## [[2]]$ability
## [[2]]$ability$url
## [1] "https://pokeapi.co/api/v2/ability/65/"
##
## [[2]]$ability$name
## [1] "overgrow"To build a Pokemon dataset for analysis, you’d need to distill the data returned from the API into a useful format; iterate across all pokemon; and combine the results.
Other materials
- A recent short course presented similar topics to those above; a GitHub repo for the course is here
- A lot of NYC data is public; this is a good place to start looking for interesting data
- There are some cool projects based on scraped data; the RStudio community collected some here
- Check out the R file used to create the
starwarsdataset (in thetidyverse) using the Star Wars API (from the maker of the Pokemon API).
The code that I produced working examples in lecture is here.