Strings and factors
Most of the tools we examined in Data Wrangling I were general purpose things – what tidy data is, using dplyr and tidyr for manipulation of data tables. Two variable types, strings and factors, present enough challenges to examine in some detail. Now might also be a good time to read up on the history of strings and factors in R!
This is the second module in the Data Wrangling II topic; the relevant slack channel is here.
Example
I’ll write code for today’s content in a new R Markdown document called strings_and_factors.Rmd, and put it in the same directory / GitHub repo as reading_data.Rmd. I’m also going to load the usual packages, as well as stringr and forcats.
library(tidyverse)
library(janitor)
library(haven)
library(rvest)
library(stringr)
library(forcats)
theme_set(theme_bw())
theme_update(legend.position = "bottom")PULSE data
We’ll start by revisiting a dataset we’ve seen a few times now. In tidy data we spent some time tidying the PULSE dataset; as part of that we used tools available to us at the time, but now we have some better tools.
The code below updates the data processing pipeline for the PULSE data using strignr and forcats. The result is the same, and the differences are pretty small, but this is a bit cleaner.
pulse_data = read_sas("./data/public_pulse_data.sas7bdat") %>%
clean_names() %>%
gather(key = visit, value = bdi, bdi_score_bl:bdi_score_12m) %>%
mutate(visit = str_replace(visit, "bdiscore_", ""),
visit = str_replace(visit, "bl", "00m"),
visit = fct_relevel(visit, str_c(c("00", "01", "06", "12"), "m"))) %>%
arrange(id, visit)
## Warning: attributes are not identical across measure variables;
## they will be dropped
## Warning: Unknown levels in `f`: 00m, 01m, 06m, 12m
print(pulse_data, n = 12)
## # A tibble: 4,348 x 5
## id age sex visit bdi
## <dbl> <dbl> <chr> <fct> <dbl>
## 1 10003 48.0 male bdi_score_00m 7
## 2 10003 48.0 male bdi_score_01m 1
## 3 10003 48.0 male bdi_score_06m 2
## 4 10003 48.0 male bdi_score_12m 0
## 5 10015 72.5 male bdi_score_00m 6
## 6 10015 72.5 male bdi_score_01m NA
## 7 10015 72.5 male bdi_score_06m NA
## 8 10015 72.5 male bdi_score_12m NA
## 9 10022 58.5 male bdi_score_00m 14
## 10 10022 58.5 male bdi_score_01m 3
## 11 10022 58.5 male bdi_score_06m 8
## 12 10022 58.5 male bdi_score_12m NA
## # ... with 4,336 more rowsNSDUH
Next we’ll revisit the table scraped from the National Survey on Drug Use and Health data on this page. In reading data from the web, we loaded this data using the code below, but noted it wasn’t tidy.
url = "http://samhda.s3-us-gov-west-1.amazonaws.com/s3fs-public/field-uploads/2k15StateFiles/NSDUHsaeShortTermCHG2015.htm"
drug_use_xml = read_html(url)
table_marj = (drug_use_xml %>% html_nodes(css = "table"))[[1]] %>%
html_table() %>%
.[-1,] %>%
as_tibble()There are a few steps we need to implement to tidy these data. For now I’m not interested in the p-values (I’d rather just see the data); we also have age groups and year ranges in column titles, both of which are, in fact, variables. Lastly, the table includes letters as superscripts next to table entries; if we only want the percents we’ll need to strip these out.
data_marj =
table_marj %>%
select(-contains("P Value")) %>%
gather(key = key, value = percent, -State) %>%
separate(key, into = c("age", "year"), sep = "\\(") %>%
mutate(year = str_sub(year, 1, -2),
percent = str_replace(percent, "[a-z]", ""),
percent = as.numeric(percent)) %>%
filter(!(State %in% c("Total U.S.", "Northeast", "Midwest", "South", "West")))We used stringr and regular expressions a couple of times above:
- in
separate, we split age and year at the open parentheses using"\\(" - to remove character superscripts, we replaced any character using
"[a-z]"
Let’s quickly visualize these data for the 12-17 age group; to make the plot readable, we’ll treat State as a factor are reorder according to percent.
data_marj %>%
filter(age == "12-17") %>%
mutate(State = fct_reorder(State, percent)) %>%
ggplot(aes(x = State, y = percent, color = year)) +
geom_point() +
theme(axis.text.x = element_text(angle = 90, hjust = 1))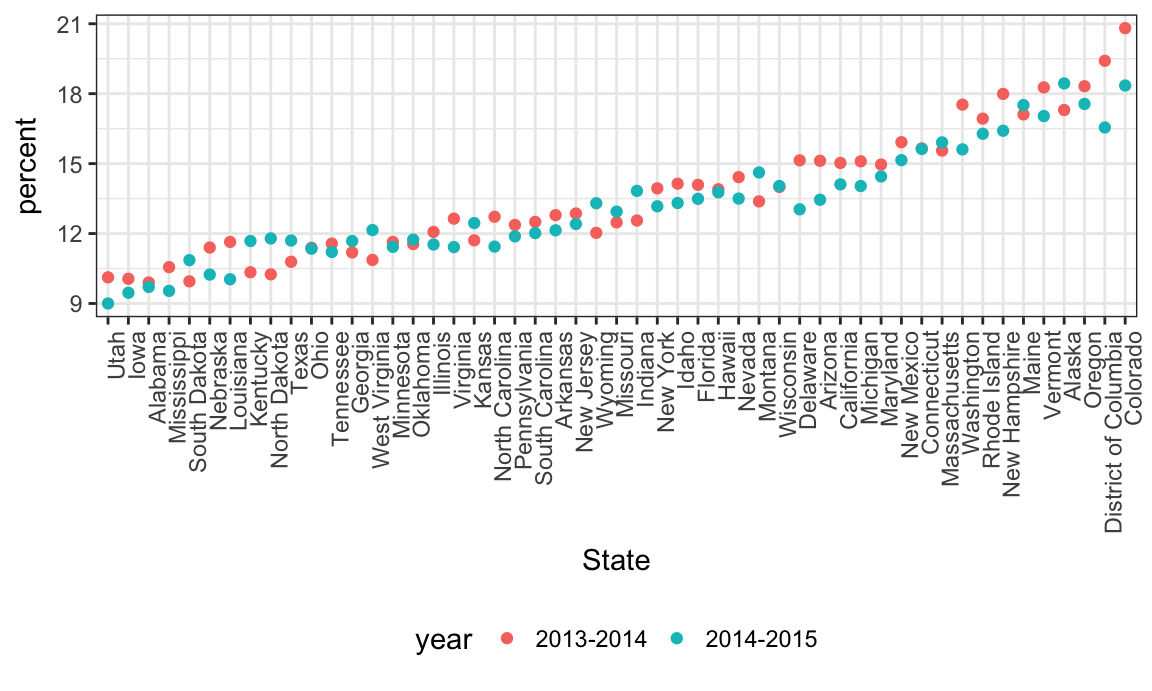
Toothbrush reviews
We were able to scrape toothbrush reviews for a single review page in reading data from the web. If we want to scrape more reviews, it’s necessary to construct the URLs for several review pages. This is possible based on an observation about the the structure of the URL for the first page of reviews – it ended with pageNumber=1 and, sure enough, changing this to pageNumber=2 will bring up the second page of reviews.
url_base = "https://www.amazon.com/Philips-Sonicare-rechargeable-toothbrush-HX6211/product-reviews/B00YAR7ZFM/ref=cm_cr_arp_d_viewopt_srt?ie=UTF8&reviewerType=all_reviews&sortBy=recent&pageNumber="
urls = str_c(url_base, 1:5)
read_html(urls[1]) %>%
html_nodes("#cm_cr-review_list .review-title") %>%
html_text()
## [1] "Five Stars"
## [2] "The BEST Toothbrush"
## [3] "Five Stars"
## [4] "It doesn't clean as good as the round spinning brush"
## [5] "Five Stars"
## [6] "One Star"
## [7] "Ni-MH battery, NOT LITHIUM!"
## [8] "I wasted my money"
## [9] "but still works fine. Great for the price"
## [10] "Four Stars"
read_html(urls[2]) %>%
html_nodes("#cm_cr-review_list .review-title") %>%
html_text()
## [1] "Still working perfectly nearly two years later"
## [2] "Five Stars"
## [3] "Significant difference in clean teeth/gums"
## [4] "Five Stars"
## [5] "Five Stars"
## [6] "A good buy"
## [7] "Works great, love it"
## [8] "One Star"
## [9] "... each of the members in my household and we love them."
## [10] "Bad battery"It would get really tedious to try to scrape all (5000+) reviews by copying code over and over; before long we’ll learn about writing R functions to reuse code and make this much easier!
Biostat Methods I Example
The following dataset was used as an example in Biostat Methods on ANOVA. First we’ll recreate the dataset.
ins1 = c(1.53, 1.61, 3.75, 2.89, 3.26)
ins2 = c(3.15, 3.96, 3.59, 1.89, 1.45, 1.56)
ins3 = c(3.89, 3.68, 5.70, 5.62, 5.79, 5.33)
ins4 = c(8.18, 5.64, 7.36, 5.33, 8.82, 5.26, 7.10)
ins5 = c(5.86, 5.46, 5.69, 6.49, 7.81, 9.03, 7.49, 8.98)
bmi_data = data_frame(
insulin = c(ins1, ins2, ins3, ins4, ins5),
ind = c(rep(1, length(ins1)),
rep(2, length(ins2)),
rep(3, length(ins3)),
rep(4, length(ins4)),
rep(5, length(ins5)))
)Next we’ll convert ind to a factor variable with informative levels, make a quick plot, and fit a linear model using ind as a predictor and insulin as the outcome.
bmi_data = bmi_data %>%
mutate(ind = factor(ind),
ind = fct_recode(ind,
"level_1" = "1",
"level_2" = "2",
"level_3" = "3",
"level_4" = "4",
"level_5" = "5")
)
bmi_data %>%
ggplot(aes(x = ind, y = insulin)) + geom_boxplot()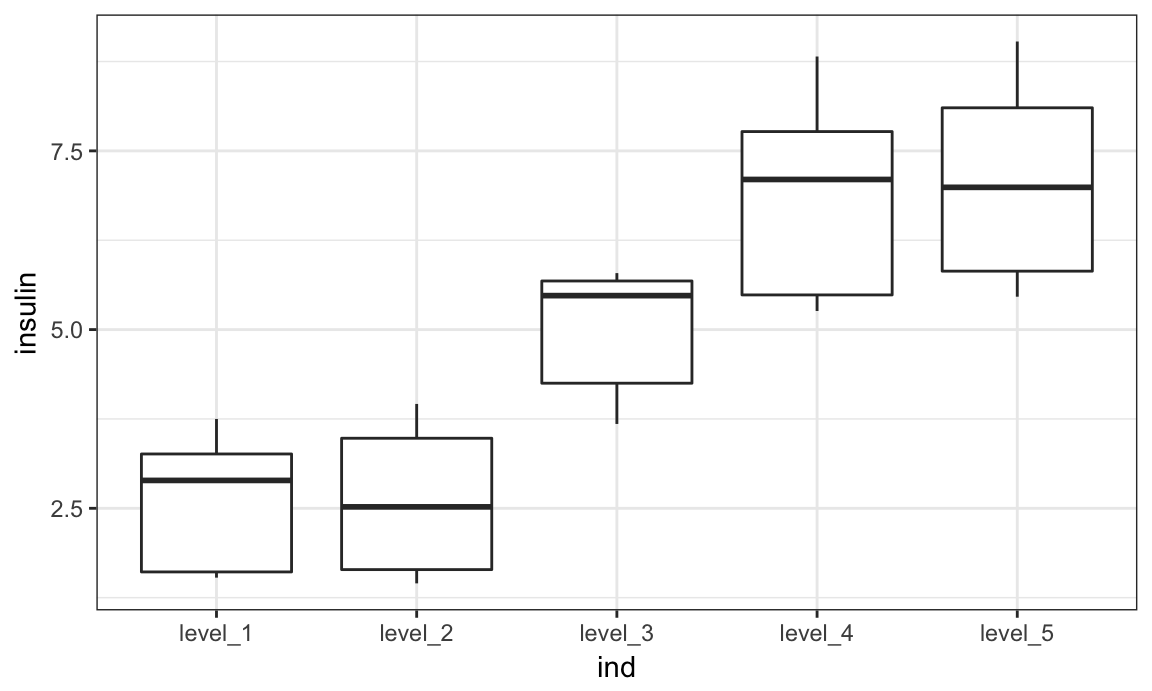
bmi_data %>%
lm(insulin ~ ind, data = .) %>%
broom::tidy() %>%
select(-std.error, -statistic) %>%
knitr::kable(digits = 3)| term | estimate | p.value |
|---|---|---|
| (Intercept) | 2.608 | 0.000 |
| indlevel_2 | -0.008 | 0.992 |
| indlevel_3 | 2.394 | 0.004 |
| indlevel_4 | 4.205 | 0.000 |
| indlevel_5 | 4.493 | 0.000 |
As it is, level_1 is used as the reference group in the regression model. That might make sense, and it might not – if the factor takes on different values you may want something else as the reference.
bmi_data = bmi_data %>%
mutate(ind = fct_recode(ind,
"very_low" = "level_1",
"low" = "level_2",
"neutral" = "level_3",
"high" = "level_4",
"very_high" = "level_5"),
ind = fct_relevel(ind, "neutral"))
bmi_data %>%
lm(insulin ~ ind, data = .) %>%
broom::tidy() %>%
select(-std.error, -statistic) %>%
knitr::kable(digits = 3)| term | estimate | p.value |
|---|---|---|
| (Intercept) | 5.002 | 0.000 |
| indvery_low | -2.394 | 0.004 |
| indlow | -2.402 | 0.002 |
| indhigh | 1.811 | 0.014 |
| indvery_high | 2.100 | 0.004 |
Shifting the reference category also changes our plot.
bmi_data %>%
ggplot(aes(x = ind, y = insulin)) + geom_boxplot()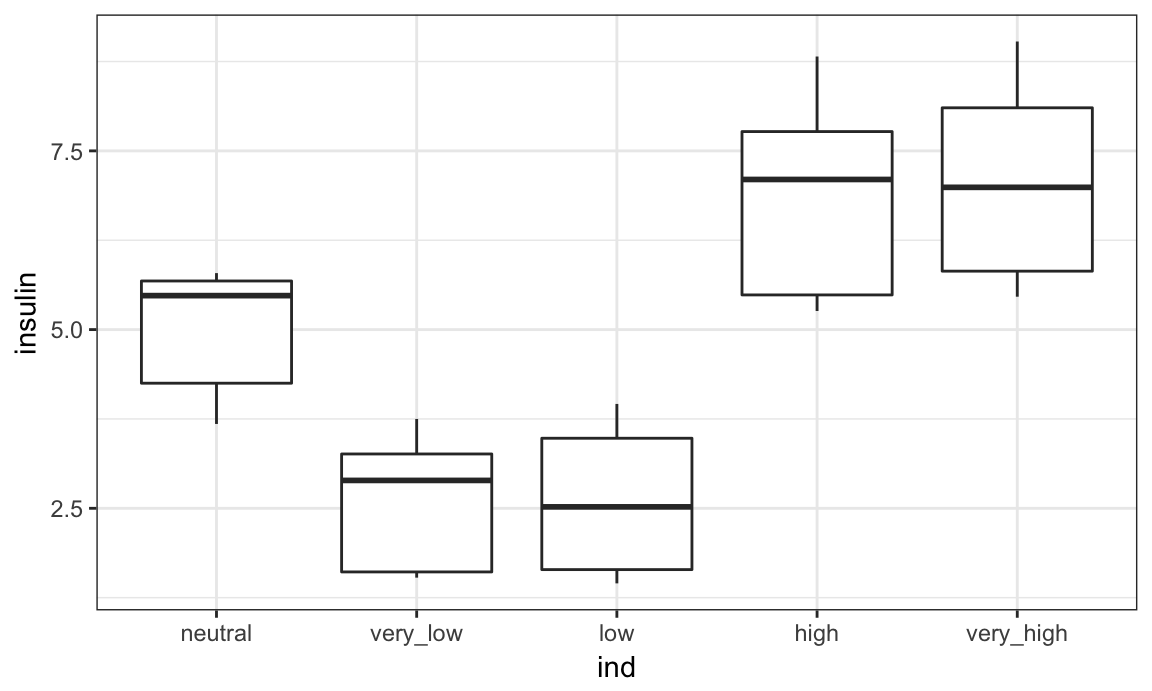
In the above example, neutral makes sense as a reference group but might not give the ordering you want for visualizations. That’s fine – you can use what you need when you need it!
Restaurant inspections
As a final example of strings and factors, we’ll take a look at the NYC Restuarant Inspections data. Although we won’t talk about this in detail now, it’s worth mentioning that these data were collected using the NYC Open Data API; code is available with the data description.
First we’ll import the data and take a look.
nyc_inspections = read_csv("./data/DOHMH_New_York_City_Restaurant_Inspection_Results.csv.gz",
col_types = cols(building = col_character()),
na = c("NA", "N/A"))
nyc_inspections %>%
group_by(boro, grade) %>%
summarize(n = n()) %>%
spread(key = grade, value = n)
## # A tibble: 6 x 8
## # Groups: boro [6]
## boro A B C `Not Yet Graded` P Z `<NA>`
## <chr> <int> <int> <int> <int> <int> <int> <int>
## 1 BRONX 13688 2801 701 200 163 351 16833
## 2 BROOKLYN 37449 6651 1684 702 416 977 51930
## 3 MANHATTAN 61608 10532 2689 765 508 1237 80615
## 4 Missing 4 NA NA NA NA NA 13
## 5 QUEENS 35952 6492 1593 604 331 913 45816
## 6 STATEN ISLAND 5215 933 207 85 47 149 6730To simplify things, I’ll remove inspections with scores other than A, B, or C, and also remove the restaurants with missing boro information. I’ll also clean up boro names a bit.
nyc_inspections =
nyc_inspections %>%
filter(grade %in% c("A", "B", "C"), boro != "Missing") %>%
mutate(boro = str_to_title(boro))Let’s focus only on pizza places for now, and re-examine grades by boro
nyc_inspections %>%
filter(str_detect(dba, "Pizza")) %>%
group_by(boro, grade) %>%
summarize(n = n()) %>%
spread(key = grade, value = n)
## # A tibble: 5 x 3
## # Groups: boro [5]
## boro A B
## <chr> <int> <int>
## 1 Bronx 9 3
## 2 Brooklyn 6 NA
## 3 Manhattan 26 8
## 4 Queens 17 NA
## 5 Staten Island 5 NAThat doesn’t look right – for sure there are more pizza places than that! The problem is that the match in str_detect is case-sensitive until we tell it not to be:
nyc_inspections %>%
filter(str_detect(dba, regex("pizza", ignore_case = TRUE))) %>%
group_by(boro, grade) %>%
summarize(n = n()) %>%
spread(key = grade, value = n)
## # A tibble: 5 x 4
## # Groups: boro [5]
## boro A B C
## <chr> <int> <int> <int>
## 1 Bronx 1170 305 56
## 2 Brooklyn 1948 296 61
## 3 Manhattan 1983 420 76
## 4 Queens 1647 259 48
## 5 Staten Island 323 127 21The table is okay but I’d like to visualize this.
nyc_inspections %>%
filter(str_detect(dba, regex("pizza", ignore_case = TRUE))) %>%
ggplot(aes(x = boro, fill = grade)) + geom_bar()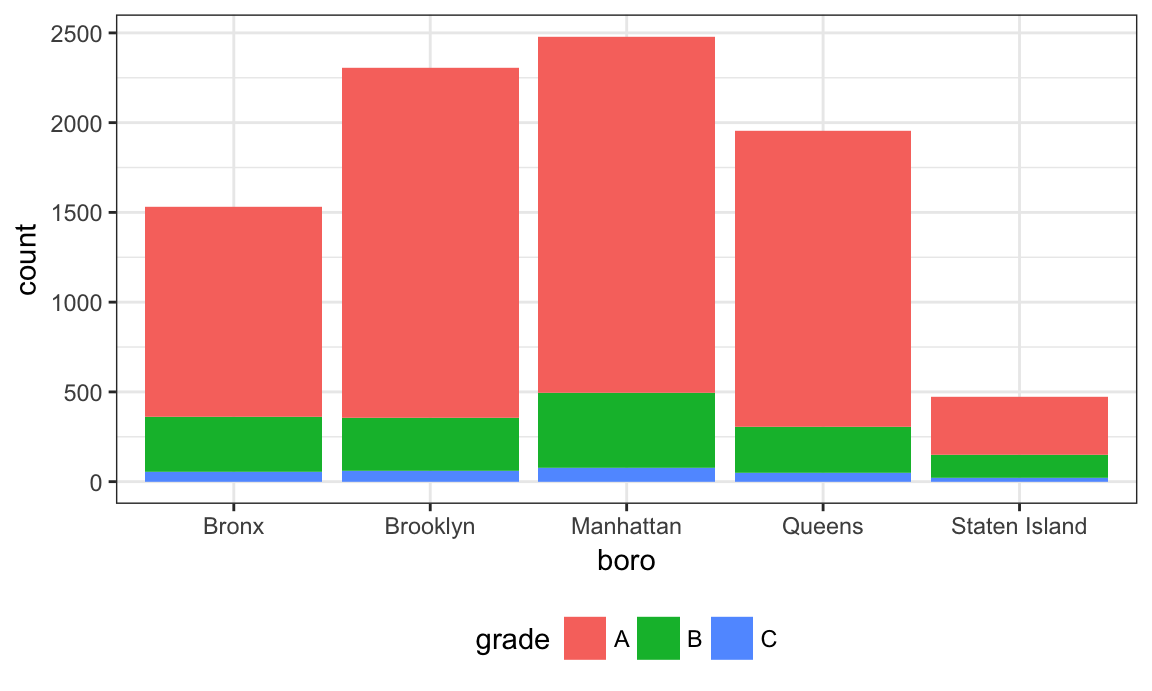
Might help to have things in a different order – maybe number of pizza places? Fortunately this can be done using fct_infreq.
nyc_inspections %>%
filter(str_detect(dba, regex("pizza", ignore_case = TRUE))) %>%
mutate(boro = fct_infreq(boro)) %>%
ggplot(aes(x = boro, fill = grade)) + geom_bar()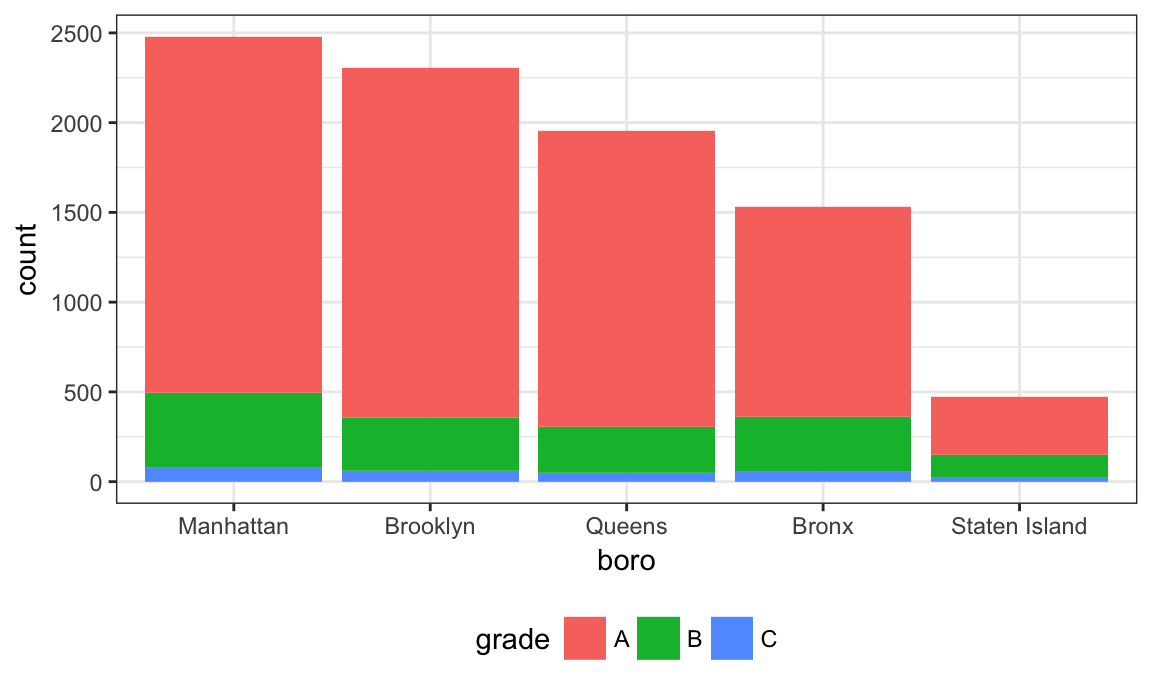
Suppose I want to rename a boro; I’ll try to do this using replace.
nyc_inspections %>%
filter(str_detect(dba, regex("pizza", ignore_case = TRUE))) %>%
mutate(boro = fct_infreq(boro),
boro = replace(boro, which(boro == "Brooklyn"), "Hipsterville")) %>%
ggplot(aes(x = boro, fill = grade)) + geom_bar()
## Warning in `[<-.factor`(`*tmp*`, list, value = "Hipsterville"): invalid
## factor level, NA generated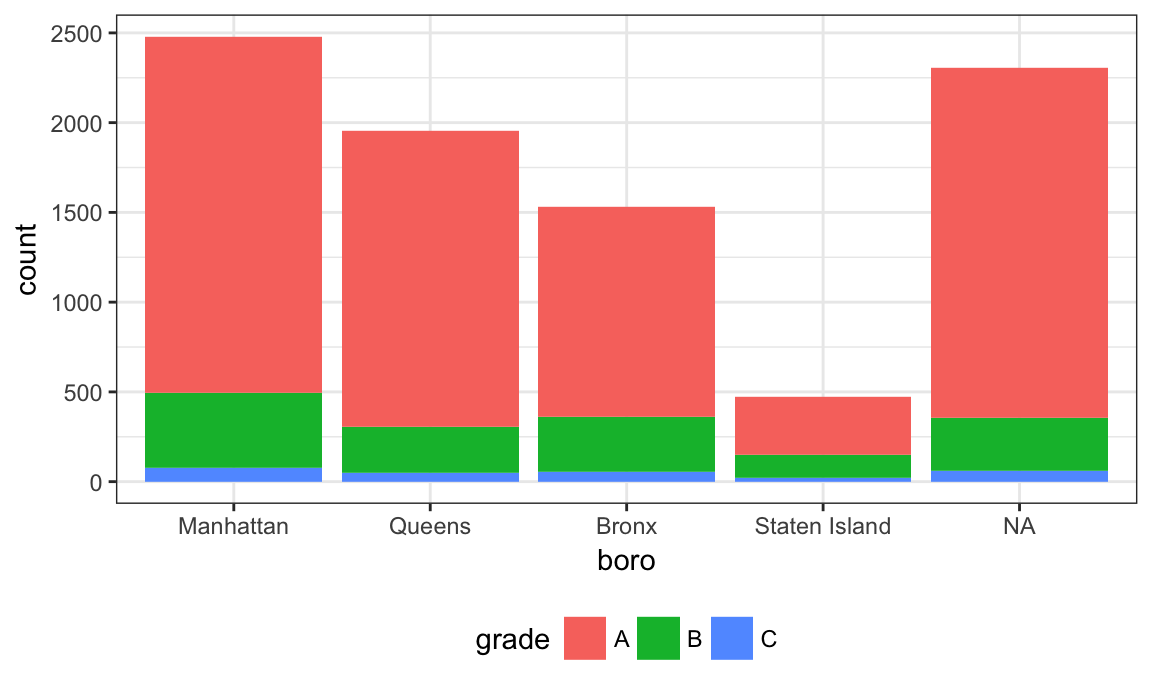
That didn’t work at all! Factors have very specific values, trying to use a value that is not an existing factor level won’t work. Fortunately there is a dedicated function for renaming factor levels:
nyc_inspections %>%
filter(str_detect(dba, regex("pizza", ignore_case = TRUE))) %>%
mutate(boro = fct_infreq(boro),
boro = fct_recode(boro, "Hipsterville" = "Brooklyn")) %>%
ggplot(aes(x = boro, fill = grade)) + geom_bar()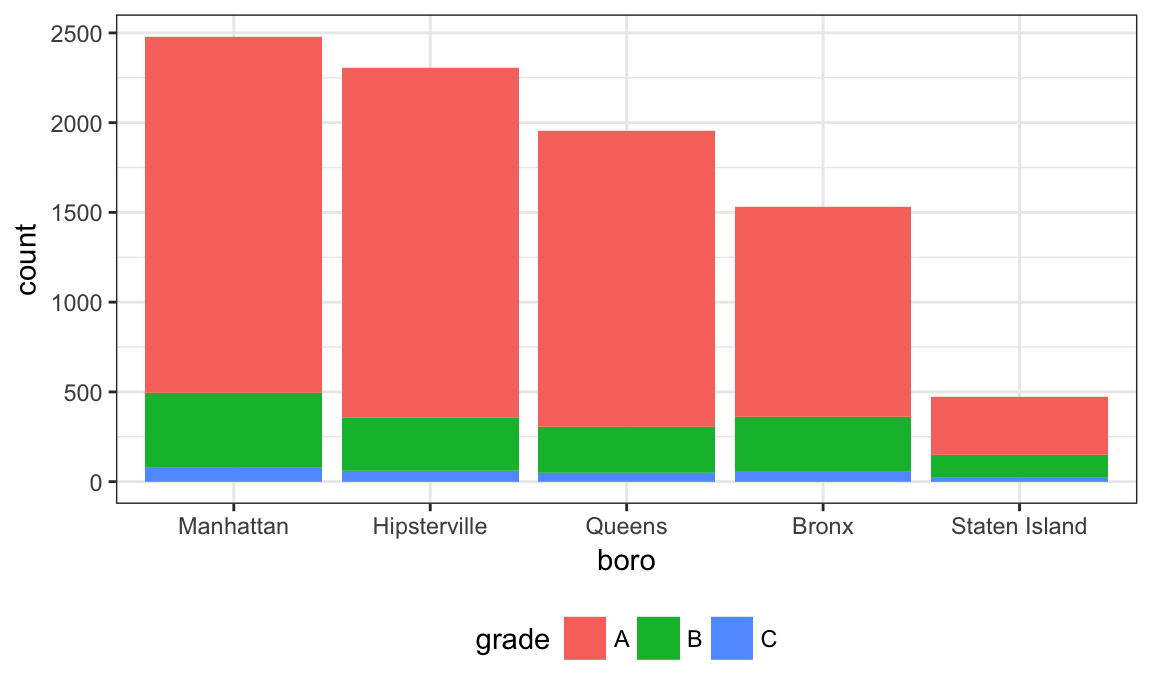
Success!
Other materials
- R for Data Science has chapters on both strings and factors
- There are useful materials by Jenny Bryan on strings and factors as well
- Dealing with text entries can be a real challenge
- Cheatsheet for stringr
The code that I produced working examples in lecture is here.